top of page
The LaBella Lab
University of North Carolina at Charlotte

Publications
No Collections Here
Sort your projects into collections. Click on "Manage Collections" to get started


Gonçalves C, Steenwyk JL, Rinker DC, Opulente DA, LaBella AL, Harrison MC, Wolters JF, Zhou X, Shen XX, Covo S, Groenewald M.
Stable hypermutators revealed by the genomic landscape of DNA repair genes among yeast species.
bioRxiv. 2025 Mar 17.
Stable hypermutators revealed by the genomic landscape of DNA repair genes among yeast species.
bioRxiv. 2025 Mar 17.


Harrison MC, Rinker DC, LaBella AL, Opulente DA, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT, Rokas A.
Machine learning identifies novel signatures of antifungal drug resistance in Saccharomycotina yeasts.
bioRxiv. 2025 May 10:2025-05.
Machine learning identifies novel signatures of antifungal drug resistance in Saccharomycotina yeasts.
bioRxiv. 2025 May 10:2025-05.

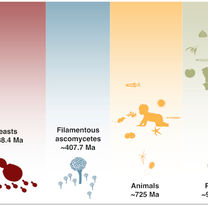
Feng B, Li Y, Xu B, Liu H, Steenwyk JL, David KT, Tian X, Gonçalves C, Opulente DA, LaBella AL, Harrison MC.
Unique trajectory of gene family evolution from genomic analysis of nearly all known species in an ancient yeast lineage.
Molecular Systems Biology. 2025 May 27:1-24.
Unique trajectory of gene family evolution from genomic analysis of nearly all known species in an ancient yeast lineage.
Molecular Systems Biology. 2025 May 27:1-24.

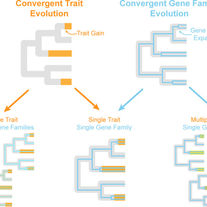
David KT, Schraiber JG, Crandall JG, Labella AL, Opulente DA, Harrison MC, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT.
Convergent expansions of keystone gene families drive metabolic innovation in Saccharomycotina yeasts.
Proceedings of the National Academy of Sciences. 2025 Jun 10;122(23):e2500165122.
Convergent expansions of keystone gene families drive metabolic innovation in Saccharomycotina yeasts.
Proceedings of the National Academy of Sciences. 2025 Jun 10;122(23):e2500165122.

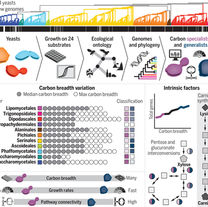
Opulente DA, LaBella AL, Harrison MC, Wolters JF, Liu C, Li Y, Kominek J, Steenwyk JL, Stoneman HR, VanDenAvond J, Miller CR.
Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts.
Science. 2024 Apr 26;384(6694):eadj4503.
Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts.
Science. 2024 Apr 26;384(6694):eadj4503.

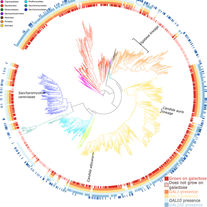
Harrison MC, Ubbelohde EJ, LaBella AL, Opulente DA, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT, Rokas A. Machine learning enables identification of an alternative yeast galactose utilization pathway. Proceedings of the National Academy of Sciences. 2024 Apr 30;121(18):e2315314121.

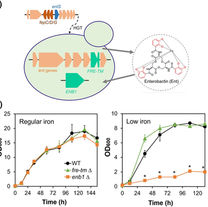
Sun L, David KT, Wolters JF, Karlen SD, Gonçalves C, Opulente DA, LaBella AL, Groenewald M, Zhou X, Shen XX, Rokas A.
Functional and evolutionary integration of a fungal gene with a bacterial operon. Molecular Biology and Evolution.
2024 Apr;41(4):msae045.
Functional and evolutionary integration of a fungal gene with a bacterial operon. Molecular Biology and Evolution.
2024 Apr;41(4):msae045.


Harrison MC, Opulente DA, Wolters JF, Shen XX, Zhou X, Groenewald M, Hittinger CT, Rokas A, LaBella AL.
Exploring Saccharomycotina Yeast Ecology Through an Ecological Ontology Framework.
Yeast. 2024 Oct;41(10):615-28.
Exploring Saccharomycotina Yeast Ecology Through an Ecological Ontology Framework.
Yeast. 2024 Oct;41(10):615-28.


David KT, Harrison MC, Opulente DA, LaBella AL, Wolters JF, Zhou X, Shen XX, Groenewald M, Pennell M, Hittinger CT, Rokas A.
Saccharomycotina yeasts defy long-standing macroecological patterns.
Proceedings of the National Academy of Sciences. 2024 Mar 5;121(10):e2316031121.
Saccharomycotina yeasts defy long-standing macroecological patterns.
Proceedings of the National Academy of Sciences. 2024 Mar 5;121(10):e2316031121.


Nalabothu RL, Fisher KJ, LaBella AL, Meyer TA, Opulente DA, Wolters JF, Rokas A, Hittinger CT.
Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts.
Molecular biology and evolution. 2023 Jun;40(6):msad111.
Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts.
Molecular biology and evolution. 2023 Jun;40(6):msad111.


Groenewald M, Hittinger CT, Bensch K, Opulente DA, Shen XX, Li Y, Liu C, LaBella AL, Zhou X, Limtong S, Jindamorakot S.
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina.
Studies in Mycology. 2023 Jun 15;105(1):1-22.
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina.
Studies in Mycology. 2023 Jun 15;105(1):1-22.

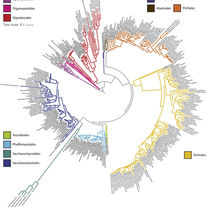
Wolters JF, LaBella AL, Opulente DA, Rokas A, Hittinger CT.
Mitochondrial genome diversity across the subphylum Saccharomycotina.
Frontiers in Microbiology. 2023 Nov 23;14:1268944.
Mitochondrial genome diversity across the subphylum Saccharomycotina.
Frontiers in Microbiology. 2023 Nov 23;14:1268944.


Harrison MC, LaBella AL, Hittinger CT, Rokas A.
The evolution of the GALactose utilization pathway in budding yeasts.
Trends in Genetics. 2022 Jan 1;38(1):97-106.
The evolution of the GALactose utilization pathway in budding yeasts.
Trends in Genetics. 2022 Jan 1;38(1):97-106.

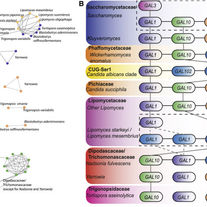
Li Y, Liu H, Steenwyk JL, LaBella AL, Harrison MC, Groenewald M, Zhou X, Shen XX, Zhao T, Hittinger CT, Rokas A.
Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum.
Current Biology. 2022 Dec 19;32(24):5335-43.
Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum.
Current Biology. 2022 Dec 19;32(24):5335-43.


Haase MA, Kominek J, Opulente DA, Shen XX, LaBella AL, Zhou X, DeVirgilio J, Hulfachor AB, Kurtzman CP, Rokas A, Hittinger CT.
Repeated horizontal gene transfer of GAL actose metabolism genes violates
Dollo’s law of irreversible loss. Genetics. 2021 Feb 1;217(2):iyaa012.
Repeated horizontal gene transfer of GAL actose metabolism genes violates
Dollo’s law of irreversible loss. Genetics. 2021 Feb 1;217(2):iyaa012.

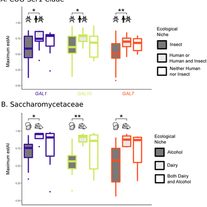
LaBella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A.
Signatures of optimal codon usage in metabolic genes inform budding yeast ecology.
PLoS biology. 2021 Apr 19;19(4):e3001185.
Signatures of optimal codon usage in metabolic genes inform budding yeast ecology.
PLoS biology. 2021 Apr 19;19(4):e3001185.


Steenwyk JL, Opulente DA, Kominek J, Shen XX, Zhou X, Labella AL, Bradley NP, Eichman BF, Čadež N, Libkind D, DeVirgilio J.
Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts.
PLoS Biology. 2019 May 21;17(5):e3000255.
Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts.
PLoS Biology. 2019 May 21;17(5):e3000255.
bottom of page