top of page
The LaBella Lab
University of North Carolina at Charlotte

Publications
No Collections Here
Sort your projects into collections. Click on "Manage Collections" to get started


Lee S, West C, Opulente DA, Harrison MC, Wolters JF, Shen XX, Zhou X, Groenewald M, Hittinger CT, Rokas A, LaBella AL.
Genomic factors limiting the diversity of Saccharomycotina plant pathogens.
G3 (Bethesda). 2025 Aug 12:jkaf184.
Genomic factors limiting the diversity of Saccharomycotina plant pathogens.
G3 (Bethesda). 2025 Aug 12:jkaf184.


Feng B, Li Y, Xu B, Liu H, Steenwyk JL, David KT, Tian X, Gonçalves C, Opulente DA, LaBella AL, Harrison MC.
Unique trajectory of gene family evolution from genomic analysis of nearly all known species in an ancient yeast lineage.
Molecular Systems Biology. 2025 May 27:1-24.
Unique trajectory of gene family evolution from genomic analysis of nearly all known species in an ancient yeast lineage.
Molecular Systems Biology. 2025 May 27:1-24.

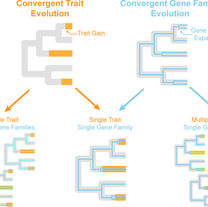
David KT, Schraiber JG, Crandall JG, Labella AL, Opulente DA, Harrison MC, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT.
Convergent expansions of keystone gene families drive metabolic innovation in Saccharomycotina yeasts.
Proceedings of the National Academy of Sciences. 2025 Jun 10;122(23):e2500165122.
Convergent expansions of keystone gene families drive metabolic innovation in Saccharomycotina yeasts.
Proceedings of the National Academy of Sciences. 2025 Jun 10;122(23):e2500165122.


Harrison MC, Rinker DC, LaBella AL, Opulente DA, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT, Rokas A.
Machine learning identifies novel signatures of antifungal drug resistance in Saccharomycotina yeasts.
bioRxiv. 2025 May 10:2025-05.
Machine learning identifies novel signatures of antifungal drug resistance in Saccharomycotina yeasts.
bioRxiv. 2025 May 10:2025-05.


Gonçalves C, Steenwyk JL, Rinker DC, Opulente DA, LaBella AL, Harrison MC, Wolters JF, Zhou X, Shen XX, Covo S, Groenewald M.
Stable hypermutators revealed by the genomic landscape of DNA repair genes among yeast species.
bioRxiv. 2025 Mar 17.
Stable hypermutators revealed by the genomic landscape of DNA repair genes among yeast species.
bioRxiv. 2025 Mar 17.

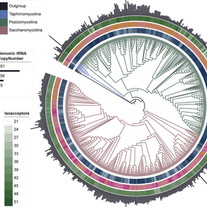
LaBella AL. A Symphony of Roles for Codon Usage in Fungal Genomics. InFungal Genomics 2024 Dec 23 (pp. 75-100). Cham: Springer Nature Switzerland.


Zavala B, Dineen L, Fisher KJ, Opulente DA, Harrison MC, Wolters JF, Shen XX, Zhou X, Groenewald M, Hittinger CT, Rokas A.
Genomic factors shaping codon usage across the Saccharomycotina subphylum.
G3: Genes, Genomes, Genetics. 2024 Nov;14(11):jkae207.
Genomic factors shaping codon usage across the Saccharomycotina subphylum.
G3: Genes, Genomes, Genetics. 2024 Nov;14(11):jkae207.


Opulente DA, LaBella AL, Harrison MC, Wolters JF, Liu C, Li Y, Kominek J, Steenwyk JL, Stoneman HR, VanDenAvond J, Miller CR.
Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts.
Science. 2024 Apr 26;384(6694):eadj4503.
Genomic factors shape carbon and nitrogen metabolic niche breadth across Saccharomycotina yeasts.
Science. 2024 Apr 26;384(6694):eadj4503.


Solé-Navais P, Juodakis J, Ytterberg K, Wu X, Bradfield JP, Vaudel M, LaBella AL, Helgeland Ø, Flatley C, Geller F, Finel M. Genome-wide analyses of neonatal jaundice reveal a marked departure from adult bilirubin metabolism. Nature communications. 2024 Aug 30;15(1):7550.


Harrison MC, Opulente DA, Wolters JF, Shen XX, Zhou X, Groenewald M, Hittinger CT, Rokas A, LaBella AL.
Exploring Saccharomycotina Yeast Ecology Through an Ecological Ontology Framework.
Yeast. 2024 Oct;41(10):615-28.
Exploring Saccharomycotina Yeast Ecology Through an Ecological Ontology Framework.
Yeast. 2024 Oct;41(10):615-28.


Gonçalves C, Harrison MC, Steenwyk JL, Opulente DA, LaBella AL, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT, Rokas A.
Diverse signatures of convergent evolution in cactus-associated yeasts.
Plos Biology. 2024 Sep 23;22(9):e3002832.
Diverse signatures of convergent evolution in cactus-associated yeasts.
Plos Biology. 2024 Sep 23;22(9):e3002832.

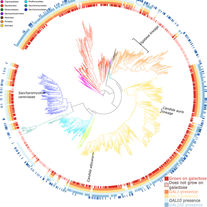
Harrison MC, Ubbelohde EJ, LaBella AL, Opulente DA, Wolters JF, Zhou X, Shen XX, Groenewald M, Hittinger CT, Rokas A. Machine learning enables identification of an alternative yeast galactose utilization pathway. Proceedings of the National Academy of Sciences. 2024 Apr 30;121(18):e2315314121.


David KT, Harrison MC, Opulente DA, LaBella AL, Wolters JF, Zhou X, Shen XX, Groenewald M, Pennell M, Hittinger CT, Rokas A.
Saccharomycotina yeasts defy long-standing macroecological patterns.
Proceedings of the National Academy of Sciences. 2024 Mar 5;121(10):e2316031121.
Saccharomycotina yeasts defy long-standing macroecological patterns.
Proceedings of the National Academy of Sciences. 2024 Mar 5;121(10):e2316031121.


Sun L, David KT, Wolters JF, Karlen SD, Gonçalves C, Opulente DA, LaBella AL, Groenewald M, Zhou X, Shen XX, Rokas A.
Functional and evolutionary integration of a fungal gene with a bacterial operon. Molecular Biology and Evolution.
2024 Apr;41(4):msae045.
Functional and evolutionary integration of a fungal gene with a bacterial operon. Molecular Biology and Evolution.
2024 Apr;41(4):msae045.


Abraham A, LaBella AL, Benton ML, Rokas A, Capra JA.
GSEL: a fast, flexible python package for detecting signatures of diverse evolutionary forces on genomic regions.
Bioinformatics. 2023 Jan 1;39(1):btad037.
GSEL: a fast, flexible python package for detecting signatures of diverse evolutionary forces on genomic regions.
Bioinformatics. 2023 Jan 1;39(1):btad037.


Groenewald M, Hittinger CT, Bensch K, Opulente DA, Shen XX, Li Y, Liu C, LaBella AL, Zhou X, Limtong S, Jindamorakot S.
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina.
Studies in Mycology. 2023 Jun 15;105(1):1-22.
A genome-informed higher rank classification of the biotechnologically important fungal subphylum Saccharomycotina.
Studies in Mycology. 2023 Jun 15;105(1):1-22.


Solé-Navais P, Flatley C, Steinthorsdottir V, Vaudel M, Juodakis J, Chen J, Laisk T, LaBella AL, Westergaard D, Bacelis J, Brumpton B.
Genetic effects on the timing of parturition and links to fetal birth weight.
Nature Genetics. 2023 Apr;55(4):559-67.
Genetic effects on the timing of parturition and links to fetal birth weight.
Nature Genetics. 2023 Apr;55(4):559-67.


Nalabothu RL, Fisher KJ, LaBella AL, Meyer TA, Opulente DA, Wolters JF, Rokas A, Hittinger CT.
Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts.
Molecular biology and evolution. 2023 Jun;40(6):msad111.
Codon optimization improves the prediction of xylose metabolism from gene content in budding yeasts.
Molecular biology and evolution. 2023 Jun;40(6):msad111.


Wolters JF, LaBella AL, Opulente DA, Rokas A, Hittinger CT.
Mitochondrial genome diversity across the subphylum Saccharomycotina.
Frontiers in Microbiology. 2023 Nov 23;14:1268944.
Mitochondrial genome diversity across the subphylum Saccharomycotina.
Frontiers in Microbiology. 2023 Nov 23;14:1268944.

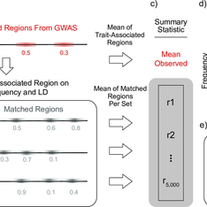
Abraham A, LaBella AL, Capra JA, Rokas A.
Mosaic patterns of selection in genomic regions associated with diverse human traits.
PLoS genetics. 2022 Nov 7;18(11):e1010494.
Mosaic patterns of selection in genomic regions associated with diverse human traits.
PLoS genetics. 2022 Nov 7;18(11):e1010494.


Harrison MC, LaBella AL, Hittinger CT, Rokas A.
The evolution of the GALactose utilization pathway in budding yeasts.
Trends in Genetics. 2022 Jan 1;38(1):97-106.
The evolution of the GALactose utilization pathway in budding yeasts.
Trends in Genetics. 2022 Jan 1;38(1):97-106.


Steenwyk JL, Buida III TJ, Gonçalves C, Goltz DC, Morales G, Mead ME, LaBella AL, Chavez CM, Schmitz JE, Hadjifrangiskou M, Li Y.
BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data.
Genetics. 2022 Jul 1;221(3):iyac079.
BioKIT: a versatile toolkit for processing and analyzing diverse types of sequence data.
Genetics. 2022 Jul 1;221(3):iyac079.

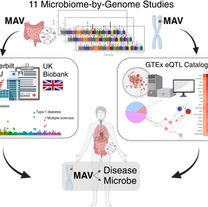
Markowitz RH, LaBella AL, Shi M, Rokas A, Capra JA, Ferguson JF, Mosley JD, Bordenstein SR.
Microbiome-associated human genetic variants impact phenome-wide disease risk.
Proceedings of the National Academy of Sciences. 2022 Jun 28;119(26):e2200551119.
Microbiome-associated human genetic variants impact phenome-wide disease risk.
Proceedings of the National Academy of Sciences. 2022 Jun 28;119(26):e2200551119.

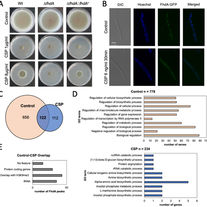
Colabardini AC, Van Rhijn N, LaBella AL, Valero C, Dineen L, Rokas A, Goldman GH.
Aspergillus fumigatus FhdA transcription factor is important for mitochondrial activity and codon usage regulation during the caspofungin paradoxical effect.
Antimicrobial Agents and Chemotherapy. 2022 Sep 20;66(9):e00701-22.
Aspergillus fumigatus FhdA transcription factor is important for mitochondrial activity and codon usage regulation during the caspofungin paradoxical effect.
Antimicrobial Agents and Chemotherapy. 2022 Sep 20;66(9):e00701-22.


Li Y, Liu H, Steenwyk JL, LaBella AL, Harrison MC, Groenewald M, Zhou X, Shen XX, Zhao T, Hittinger CT, Rokas A.
Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum.
Current Biology. 2022 Dec 19;32(24):5335-43.
Contrasting modes of macro and microsynteny evolution in a eukaryotic subphylum.
Current Biology. 2022 Dec 19;32(24):5335-43.


Hugaboom M, Hatmaker EA, LaBella AL, Rokas A.
Evolution and codon usage bias of mitochondrial and nuclear genomes in Aspergillus section Flavi.
G3. 2023 Jan 5;13(1):jkac285.
Evolution and codon usage bias of mitochondrial and nuclear genomes in Aspergillus section Flavi.
G3. 2023 Jan 5;13(1):jkac285.


LaBella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A.
Signatures of optimal codon usage in metabolic genes inform budding yeast ecology.
PLoS biology. 2021 Apr 19;19(4):e3001185.
Signatures of optimal codon usage in metabolic genes inform budding yeast ecology.
PLoS biology. 2021 Apr 19;19(4):e3001185.


Haase MA, Kominek J, Opulente DA, Shen XX, LaBella AL, Zhou X, DeVirgilio J, Hulfachor AB, Kurtzman CP, Rokas A, Hittinger CT.
Repeated horizontal gene transfer of GAL actose metabolism genes violates
Dollo’s law of irreversible loss. Genetics. 2021 Feb 1;217(2):iyaa012.
Repeated horizontal gene transfer of GAL actose metabolism genes violates
Dollo’s law of irreversible loss. Genetics. 2021 Feb 1;217(2):iyaa012.

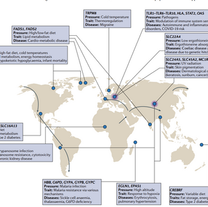
Benton ML, Abraham A, LaBella AL, Abbot P, Rokas A, Capra JA.
The influence of evolutionary history on human health and disease.
Nature Reviews Genetics. 2021 May;22(5):269-83.
The influence of evolutionary history on human health and disease.
Nature Reviews Genetics. 2021 May;22(5):269-83.


Steenwyk JL, Buida III TJ, Labella AL, Li Y, Shen XX, Rokas A.
PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data.
Bioinformatics. 2021 Aug 15;37(16):2325-31.
PhyKIT: a broadly applicable UNIX shell toolkit for processing and analyzing phylogenomic data.
Bioinformatics. 2021 Aug 15;37(16):2325-31.

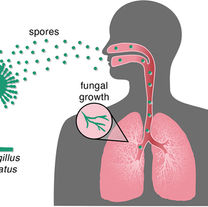
Steenwyk JL, Mead ME, de Castro PA, Valero C, Damasio A, Dos Santos RA, Labella AL, Li Y, Knowles SL, Raja HA, Oberlies NH.
Genomic and phenotypic analysis of COVID-19-associated pulmonary aspergillosis isolates of Aspergillus fumigatus.
Microbiology spectrum. 2021 Aug 31;9(1):10-128.
Genomic and phenotypic analysis of COVID-19-associated pulmonary aspergillosis isolates of Aspergillus fumigatus.
Microbiology spectrum. 2021 Aug 31;9(1):10-128.


LaBella AL, Abraham A, Pichkar Y, Fong SL, Zhang G, Muglia LJ, Abbot P, Rokas A, Capra JA.
Accounting for diverse evolutionary forces reveals mosaic patterns of selection on human preterm birth loci.
Nature Communications. 2020 Jul 24;11(1):3731.
Accounting for diverse evolutionary forces reveals mosaic patterns of selection on human preterm birth loci.
Nature Communications. 2020 Jul 24;11(1):3731.


Shen XX, Steenwyk JL, LaBella AL, Opulente DA, Zhou X, Kominek J, Li Y, Groenewald M, Hittinger CT, Rokas A.
Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota.
Science Advances. 2020 Nov 4;6(45):eabd0079.
Genome-scale phylogeny and contrasting modes of genome evolution in the fungal phylum Ascomycota.
Science Advances. 2020 Nov 4;6(45):eabd0079.

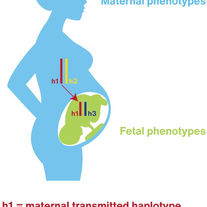
Rokas A, Mesiano S, Tamam O, LaBella A, Zhang G, Muglia L.
Developing a theoretical evolutionary framework to solve the mystery of parturition initiation.
Elife. 2020 Dec 31;9:e58343.
Developing a theoretical evolutionary framework to solve the mystery of parturition initiation.
Elife. 2020 Dec 31;9:e58343.


Labella AL, Opulente DA, Steenwyk JL, Hittinger CT, Rokas A.
Variation and selection on codon usage bias across an entire subphylum.
PLoS genetics. 2019 Jul 31;15(7):e1008304.
Variation and selection on codon usage bias across an entire subphylum.
PLoS genetics. 2019 Jul 31;15(7):e1008304.


Plouviez S, LaBella AL, Weisrock DW, von Meijenfeldt FB, Ball B, Neigel JE, Van Dover CL.
Amplicon sequencing of 42 nuclear loci supports directional gene flow between South Pacific populations of a hydrothermal vent limpet.
Ecology and Evolution. 2019 Jun;9(11):6568-80.
Amplicon sequencing of 42 nuclear loci supports directional gene flow between South Pacific populations of a hydrothermal vent limpet.
Ecology and Evolution. 2019 Jun;9(11):6568-80.


Steenwyk JL, Opulente DA, Kominek J, Shen XX, Zhou X, Labella AL, Bradley NP, Eichman BF, Čadež N, Libkind D, DeVirgilio J.
Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts.
PLoS Biology. 2019 May 21;17(5):e3000255.
Extensive loss of cell-cycle and DNA repair genes in an ancient lineage of bipolar budding yeasts.
PLoS Biology. 2019 May 21;17(5):e3000255.

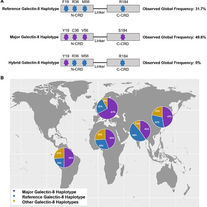
Ely ZA, Moon JM, Sliwoski GR, Sangha AK, Shen XX, Labella AL, Meiler J, Capra JA, Rokas A.
The impact of natural selection on the evolution and function of placentally expressed galectins.
Genome Biology and Evolution. 2019 Sep;11(9):2574-92.
The impact of natural selection on the evolution and function of placentally expressed galectins.
Genome Biology and Evolution. 2019 Sep;11(9):2574-92.

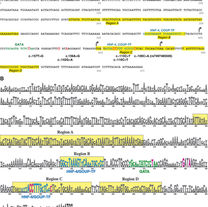
Jang YJ, LaBella AL, Feeney TP, Braverman N, Tuchman M, Morizono H, Ah Mew N, Caldovic L.
Disease�‐causing mutations in the promoter and enhancer of the ornithine transcarbamylase gene.
Human mutation. 2018 Apr;39(4):527-36.
Disease�‐causing mutations in the promoter and enhancer of the ornithine transcarbamylase gene.
Human mutation. 2018 Apr;39(4):527-36.

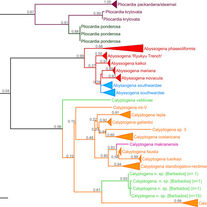
LaBella AL, Van Dover CL, Jollivet D, Cunningham CW.
Gene flow between Atlantic and Pacific Ocean basins in three lineages of deep-sea clams (Bivalvia: Vesicomyidae: Pliocardiinae) and subsequent limited gene flow within the Atlantic.
Deep Sea Research Part II: Topical Studies in Oceanography. 2017 Mar 1;137:307-17.
Gene flow between Atlantic and Pacific Ocean basins in three lineages of deep-sea clams (Bivalvia: Vesicomyidae: Pliocardiinae) and subsequent limited gene flow within the Atlantic.
Deep Sea Research Part II: Topical Studies in Oceanography. 2017 Mar 1;137:307-17.
bottom of page